|
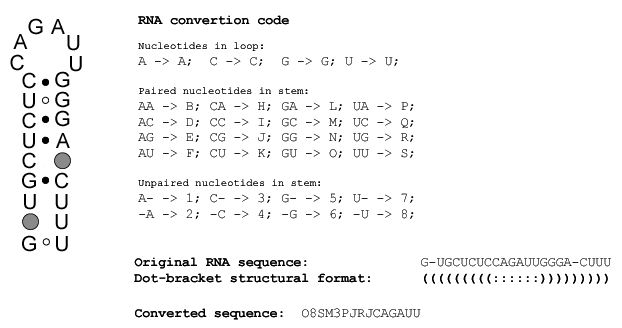
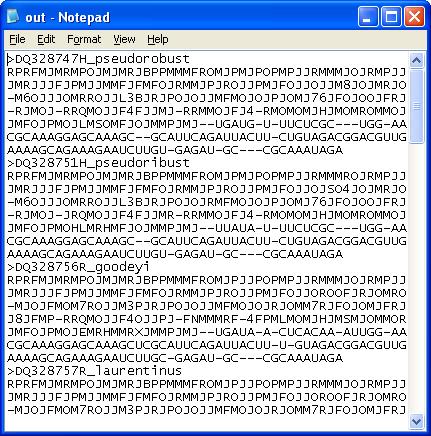
Secondary structure statistics, sequence coding,
dot-bracket structural format
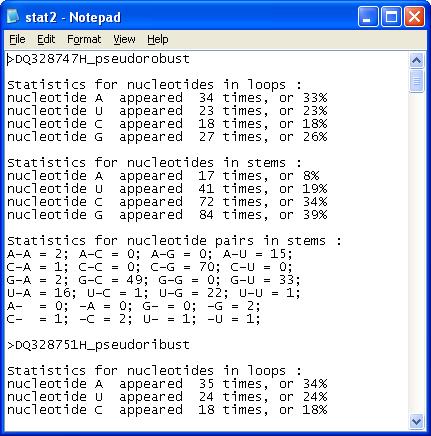
1. The program calculates and
displays: number and percentage of nucleotides in loops and stems for
each sequence; number of nucleotide pairs for each sequence; minimal,
maximal and average number of nucleotides in stems and loops; average,
minimal and maximal percentage of nucleotides in stems and loops; average,
minimal and maximal percentage of each nucleotide pair in sequence
alignment. The data is saved in the file: "stat2.txt".
Example of output
file:
|